After MM and I developed several R tools for our team (missing the whole year we kept working together), we began to look for more convenient way for the users. We were eager to build our own packages, but we didn’t know how and didn’t dig deep down. Once I found our intern was using version control while working in RStudio. The Project concept came into our view. Surprisingly, Projects are just the basic concept for building R packages. In October 2015, I tried, failed and gave up. Now the needs for building R packages become more urgent so I start again.
Prerequisites
First of all, there are two main prerequisites for building R packages:
- GNU software development tools including a C/C++ compiler; and
- LaTeX for building R manuals and vignettes.
The following steps are suitable for Windows system. If you are using other OS, please look for help on Package Development Prerequisites.
Rtools
The core software development utilities required for R package development can be obtained from the Rtools download on CRAN: https://cran.rstudio.com/bin/windows/Rtools/. After downloading and installing the version of Rtools appropriate to the version of R you are using, you should also ensure that you’ve arranged your system PATH as recommended by Rtools (you can choose to do this automatically as part of Rtools installation if you like).
MiKTeX
To build manuals and vignettes you’ll also need to install the MikTeX LaTeX distribution for Windows which you can download from here: http://miktex.org/download.
If you have installed MiKTeX before like me, it is recommended that you run the update wizard in order to get the latest updates.
Just go to Startup menu. Click All Programs. Find MikTeX then go to Maintainence and click Update.
 Then just click Next, Next and Next. If the first time going by “Selecting packages” it’s unable for you to click on “Select All”, don’t worry, finish the update process and run all the steps once again.
Then just click Next, Next and Next. If the first time going by “Selecting packages” it’s unable for you to click on “Select All”, don’t worry, finish the update process and run all the steps once again.
Packages Needed Before Starting
devtools
All check and build work is completed by package devtools.
Now find function devtools::install_github() and devtools::install_local() quite useful to install packages on GitHub or local disc.
And command devtools::document() can create or update the NAMESPACE file.
roxygen2
Package roxygen2 is needed for generating R package documentations. Will talk more in Coding section below.
packrat
Packrat is a Dependency Management System for Projects and their R Package Dependencies. Using packrat can make your R projects more:
- Isolated: Installing a new or updated package for one project won’t break your other projects, and vice versa. That’s because packrat gives each project its own private package library.
- Portable: Easily transport your projects from one computer to another, even across different platforms. Packrat makes it easy to install the packages your project depends on.
- Reproducible: Packrat records the exact package versions you depend on, and ensures those exact versions are the ones that get installed wherever you go.
Packrat is now available on CRAN, so you can install it with:
If you like to live on the bleeding edge, you can also install the development version of Packrat with:
Build A Package
Project - New Project
Start
First, we need to create a new project. On the top right corner of the RStudio window, you can find the “Project” button.
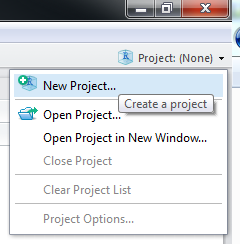 Then select “New Directory”. If you already have some R scripts but not a package yet, you can choose “Existing Directory”.
Then select “New Directory”. If you already have some R scripts but not a package yet, you can choose “Existing Directory”.
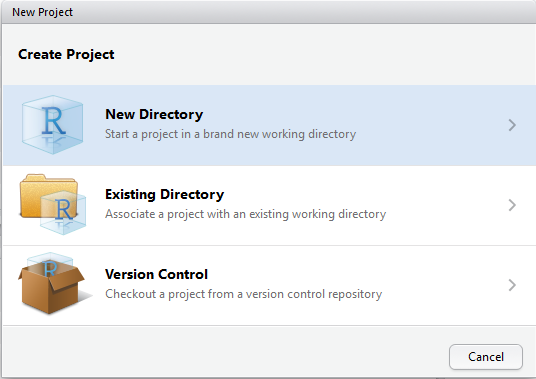 Then choose “R Package”. More exploration needed on other two choices.
Then choose “R Package”. More exploration needed on other two choices.
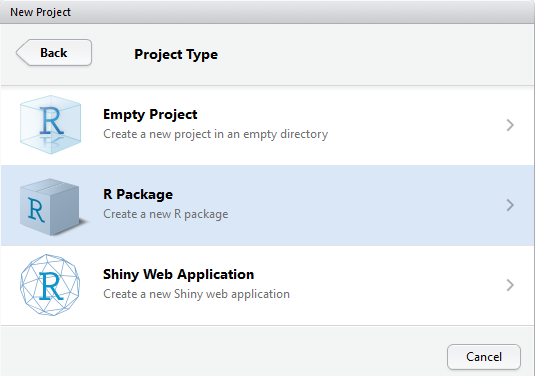 Name your package and tick “Use packrat with this project”.
Name your package and tick “Use packrat with this project”.
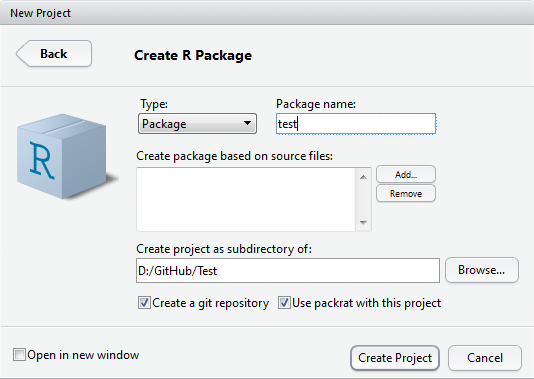 After creating a brand new package, RStudio will automatically open a new R script file “hello.R”.
After creating a brand new package, RStudio will automatically open a new R script file “hello.R”.
Under the package working directory, some files are automatically created. All the R scripts the package holds should be saved under the folder “R”.
DESCRIPTION
You can edit the package DESCRIPTION.
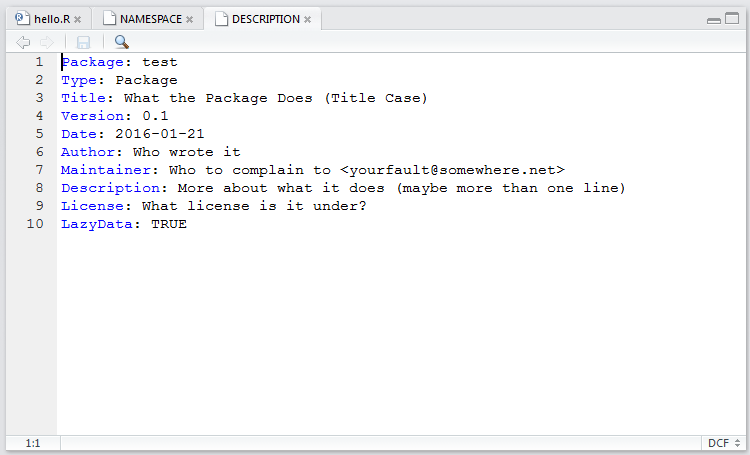 The DESCRIPTION file contains basic information about the package in the following format:
The DESCRIPTION file contains basic information about the package in the following format:
NAMESPACE
If you use roxygen2 to generate NAMESPACE automatically like me, you should not edit the NAMESPACE file by yourself. We can simply accomplish this just at the meanwhile of coding.
Dependent Packages
You can install other necessary packages in the whole process of building your package. packrat can help record the package list and their version information.
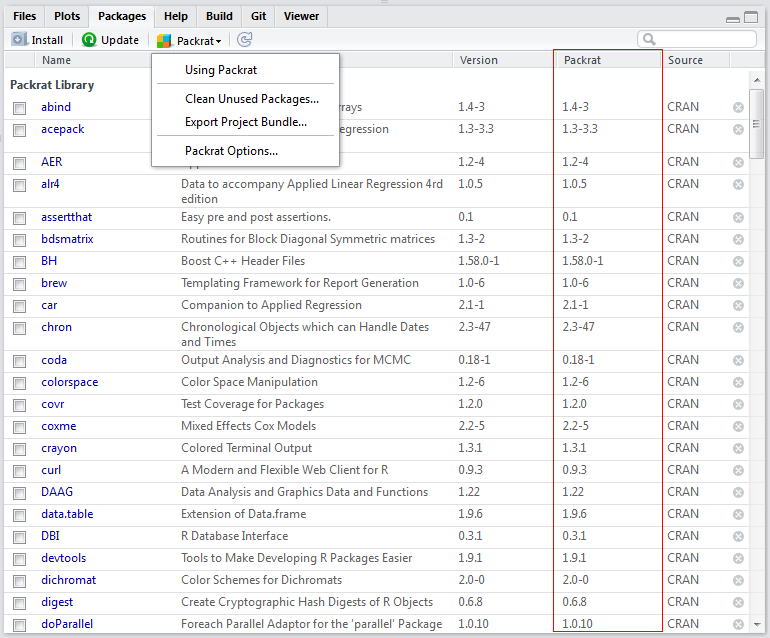
Snapshot
When you just install a packge, the “Version” and the “Source” column may have information while the “Packrat” column may be empty. The action of packrat saving information is called snapshot.
Though I choose to “Automatically snapshot local changes” in Packrat “Project Options”, I don’t see the effect after waiting for a whole night ;P
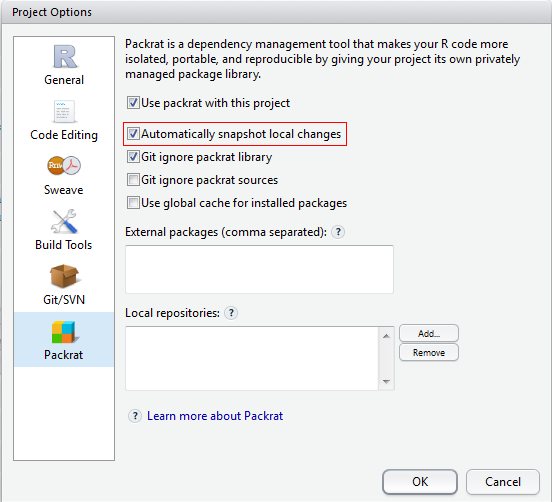 If so, you could manually snapshot by
If so, you could manually snapshot by packrat::snapshot().
Remove Packages
If you find some packages unused, you can just use remove.packages("unused package name").
Or you can just let packrat to handle it. Click “Clean Unused Packages…” and packrat begins working.
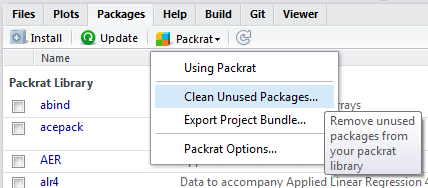
Restore
If you change the package information by mistake, you can use packrat::restore() to restore the packrat library to the latest snapshot.
Coding
Now you can just define the functions you need. What’s different is that you need to standardize the definition using roxygen2 gramma.
Here is an example which is enough for beginners. More information on Writing Package Documentation.
As you choose to use roxygen2 method as below, # Generated by roxygen2: do not edit by hand will appear at the very beginning of the NAMESPACE file.
Attention:
@param: All parameters should be mentioned and defined, or warnings would appear when checking.@export: If@exportis omitted, users are not able to call the funciton in Console.@importor@importFrom': If functions built from other packages are called inside your package, you just need to add a special comment in the format#’ @import packagenameto a file to import all functions from that package, or#’ @importFrom packagename functionnameto import one function, e.g.#’ @importFrom magrittr “%>%”
Build
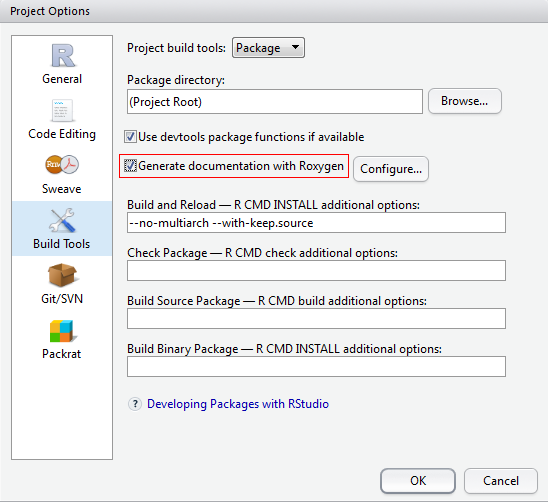
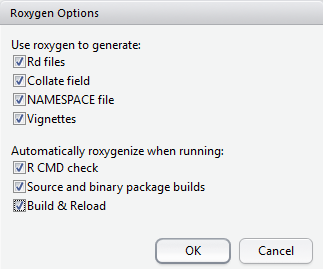
Before asking RStudio to do the “Build” work, go to “Configure Build Tools” and tick “Generate documentation with Roxygen” and all inside choices.
This can make all your #' sentences work and create .Rd files under folder man automatically. You could also not use Roxygen and create .Rd files by yourself.
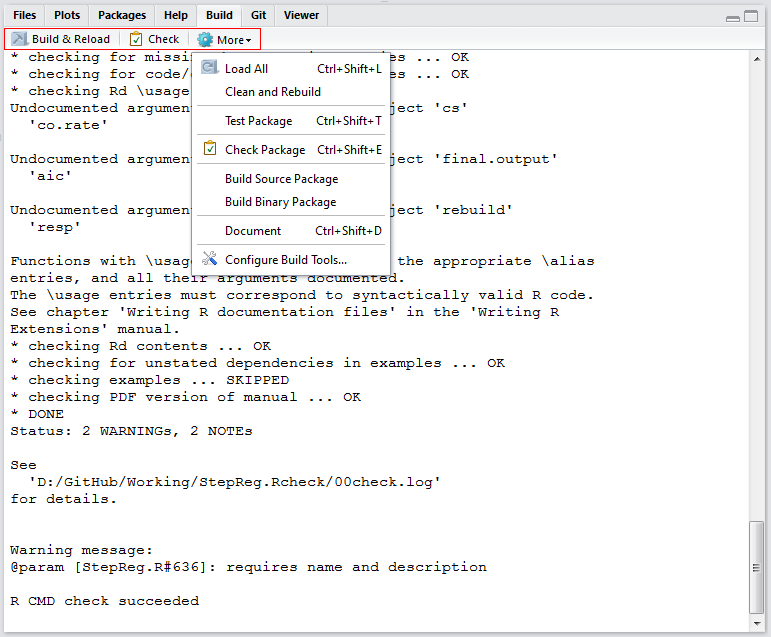
Check
RStudio will check if your package can work well and print out all the warnings, undocumented arguments, no visible global function definition, etc. After the check finishes, read the information carefully and review your code.
Build & Reload
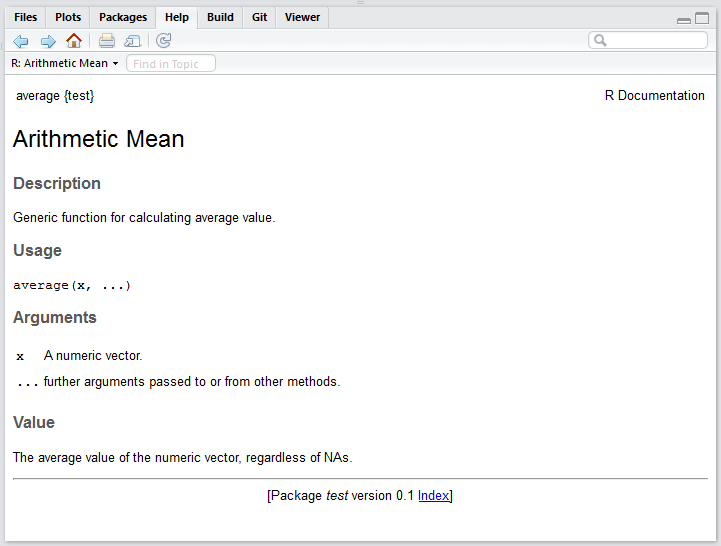
After there is no problems of the check, click “Build & Reload”. Now it’s time for you to library(YourPackage) and debug yourself.
Try ?average and see the documention built by Roxygen.
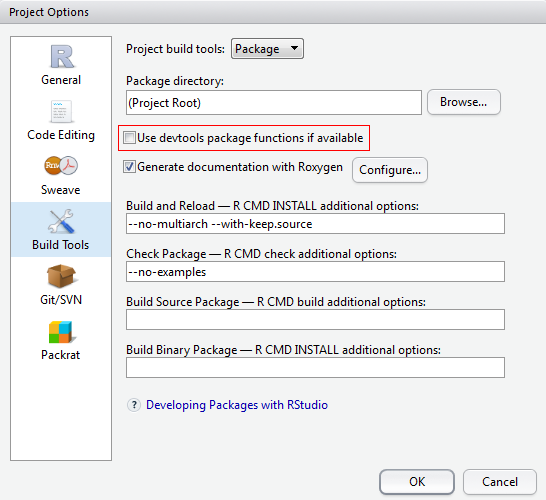
 If now error occurs as below, but you’re quite sure that the
If now error occurs as below, but you’re quite sure that the otherpackage is installed and works well.
Try unselecting the circled option in “Project Options” > “Build Tools” dialogue solves this problem, although I’m not quite clear on why:
More - Build Source Package & Build Binary Package
“Build Source Package” can output a .tar.gz file; while “Build Binary Package” can output a .zip file.
Then it’s very convenient to share your package. After the user recieve the file, they can go straight to use your packge!

